AI driven coding for life science

-
The goal of our project is to offer an environment, enabling life scientists, to perform basic bioinformatics analyses using only natural language and a brief online training.
Life scientists and programming
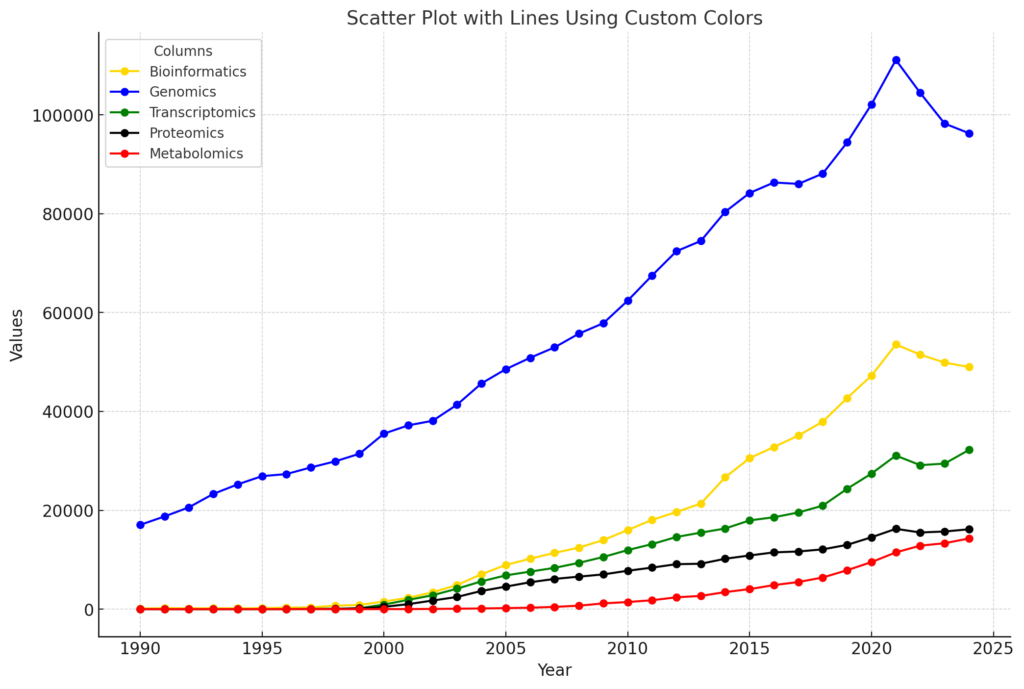
Bioinformatics has become an essential tool for managing high-throughput data. As illustrated in the figure below, bioinformatics is experiencing faster growth compared to most life science omics fields, with the exception of genomics.
Bioinformaticians frustration
It is important to note that bioinformaticians remain scarce, and many find the simplicity of routine bioinformatics analyses—such as detecting differentially expressed genes—frustrating. These tasks are relatively straightforward and, in principle, do not require highly specialized expertise but rather a life scientist trained in basic analysis techniques.
Life scientists frustration
However, learning a programming language requires a significant time investment, which poses a challenge for life scientists who already dedicate many hours daily to bench work.
Our idea
User interfaces (UIs) are not the ultimate solution for simplifying life for those without programming experience. GUIs lack generality, restrict options, and demand extensive maintenance to stay aligned with users’ evolving needs.
The advent of generative artificial intelligence, based on large language models (LLMs), presents new opportunities for programming, especially in the area of life sciences and medical research. LLMs enable programming code to be written using natural language.
Nonetheless, using LLMs requires not only a clear definition of the problem but also some familiarity with programming language concepts. Moreover, current LLM-based tools do not allow direct interaction with computational objects, such as data generated by sequencers, nor do they facilitate the evaluation of results produced by the generated code.
Our idea focuses on overcoming these limitations. The core of our concept lies in developing a software tool that enables users to interact through coherent natural language sentences, without requiring specific knowledge of a programming language. The software will execute directives written in natural language and guide researchers in defining the best analytical approach to address their problems, significantly simplifying the entire process. Our infrastructure offers unique features, ensuring reproducibility and providing a life science-friendly bioinformatics environment.
Our experience
BGcore is a Bioinformatics and Genomics facility at the University of Torino. In addition to offering wet lab and bioinformatics services, we are strongly committed to training and helping life science researchers in bioinformatics.
It is nearly 20 year that we develop tools to facilitate the access to bioinformatics analysis to life scientists building bioinformatics tools:
- oneChannelGUI (Bioinformatics 2007 Dec 15;23(24):3406-8)
- docker4seq (Bioinformatics 2018 Mar 1;34(5):871-872)
- rCASC (Gigascience 2019 Sep 1;8(9):giz105)
- CREDO (BMC Bioinformatics 2024 Mar 12;25(1):110)
Furthermore, it is more that 10 year that we run bioinformatics courses for life scientists:
- Whole transcriptome (EMBL, Heidelberg, Germany, 2012-present)
- Bulk-RNAseq and Chipseq (Duke-NUS, Singapore, 2015-2020)
- Bulk-RNAseq and single cell RNAseq (Duke-NUS, Singapore, 2023-present)
- Elixir scRNA-seq Workshop. An introduction to single cell RNA-seq data analysis (UNITO, Italy, 2022-2023).
- Elixir scRNA-seq Workshop. (UNIBA, Italy, 2025)
- Personalised data analysis courses on demand (2010-present)
WE need you! If you are a life scientist seeking to analyze your high-throughput data, you are our ideal beta-tester
We are seeking beta tester for our tool. We offer an online face-to-face training (2 hours) designed to teach you how to perform bioinformatics analyses using natural language within our computational environment. During the training, you will learn how to construct query to be converted in R or Python scripts. We will discuss with you what is your analytical goal and we will provide suggestions how to build the queries to analyse your own data.
Afterward, you will have access to our computational environment for additional 12 days. The analyses you perform will be provided both as results and as the code used to generate them. We will only request you to answer to a feed back form that will help us in improving the tool we are developing.
If you are interested to become one of our beta-tester please fill this form. You will be contacted to start the beta-testing.
We expect to have our infrastructure ready for testing in Q3 2025.
For any further information feel free to contact the reference of our project (raffaele[dot]calogero@unito[dot]it).